Our life begins with a single cell, which has the potential to develop into more than 250 different cell types in the body. Research teams from Freiburg and Milan are working to understand the still not fully understood process that ensures, at the beginning of life, a single cell eventually becomes nerve cells, skin cells, or muscle cells.
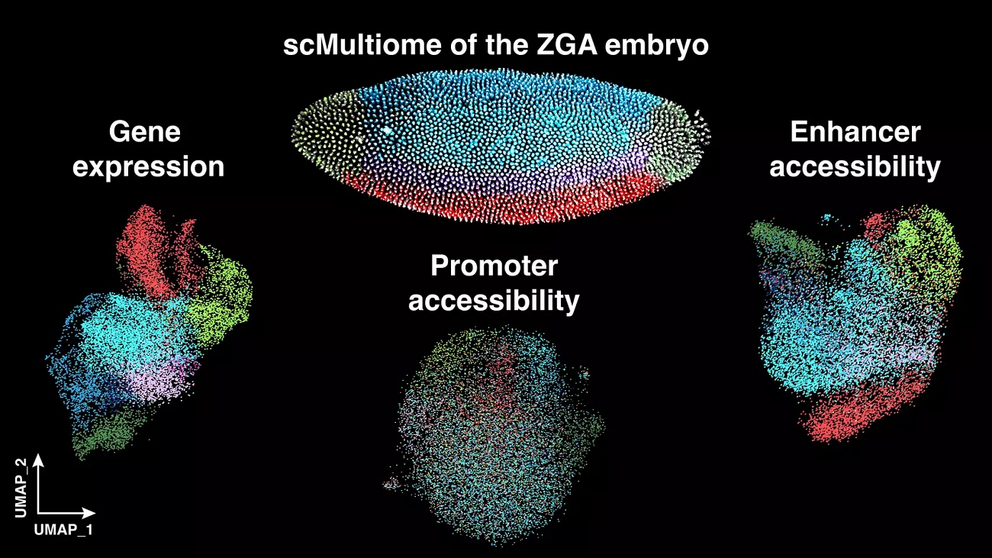
The team led by Dr. Nicola Iovino at the Max Planck Institute of Immunobiology and Epigenetics studied how epigenetic mechanisms control early cell differentiation in the embryo of the fruit fly Drosophila melanogaster. Using single-cell technologies, the researchers simultaneously analyzed the accessibility of genetic material through chromatin and the actual activity of relevant genes. Their findings reveal a finely tuned interplay between epigenetic and transcriptional processes that ensure each cell receives and maintains its proper identity, as well as the surprising discovery that chromatin accessibility and gene expression can be independently regulated during early embryonic development.
How and when do cells acquire their identity?
During zygotic genome activation (ZGA), the moment when the embryonic genome first becomes active, thousands of genes are simultaneously activated to give rise to the various cell identities, which eventually develop into specialized cell types. “In simple terms, this happens by utilizing specific regulatory regions of the genome, which act as »control centers«. They determine which histone modifications and transcription factors must bind in each cell to initiate the cell fate specification,” explains Francesco Cardamone, co-first author of the study from Freiburg. Histone modifications alter the structure of chromatin, affecting how tightly the DNA is packaged and influence gene expression. Similarly, transcription factors, which bind to the DNA, are essential for initiating gene expression.
“We found that just a few hours after fertilization, the accessibility of the genome at so-called enhancer regions is more important for determining cell identity than the accessibility at promoters,” says Nicola Iovino. “We also observed a remarkable coordination between different epigenetic mechanisms that work together to turn off the wrong genes and switch on the right ones at the appropriate time, allowing cells to develop their specific functions.”
Histone modifications tune cell fate
For example, a histone modification inherited from the mother, called H3K27me3, prevents genes that should only be active in specific cell types from being accidentally activated in other cells. This "protective function" avoids the misprogramming of cells during early embryogenesis. In contrast, the protein CBP, which sets the histone modification H3K27ac, has the opposite task: it activates transcriptional programs that ensure cells develop their proper identity. “If the modification is missing, enhancer activity and transcription come to a complete halt, causing the cells to remain trapped in an undifferentiated state, with severe consequences for development,” says co-first author Annamaria Piva from Milan.