Prof. Dr. Rolf Backofen (CIBSS-AI), Department of Computer Science (Faculty of Engineering), University of Freiburg
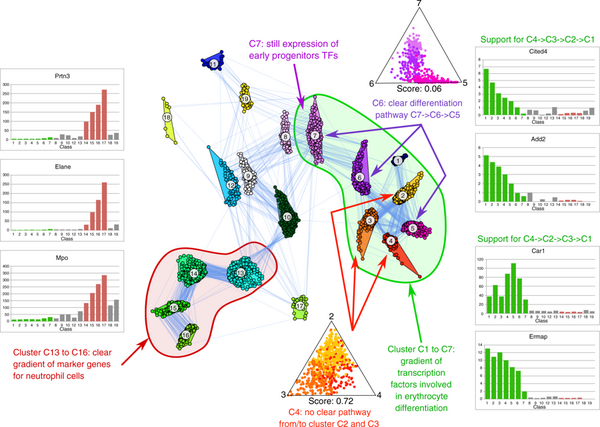
Single-Cell RNA-seq (scRNA-seq) investigates the transcriptional profile of several hundreds to a few thousand of cells and is capable of providing a genome-wide transcriptome of each single cell. scRNA-seq allows to identify cell identities, where state-of-the-art approaches for cell type classification use clustering to identify subpopulations of cells that share similar transcriptional profiles. Another task is to determine differentiation pathways and associated regulatory network. We have recently introduced GraphDPP an informed, force-based layout approach that uses a given clustering as prior information to determine differentiation pathways, thus solving both problems in parallel. Associated gene regulatory networks can be found by determining coexpression modules, however, this requires the combination of several scRNA-seq data sets. Combination can be done by aligning cells ordered by pseudotime, which, however, ignores cell type information determined from the clustering.